Antimicrobial susceptibility patterns and molecular phylogenetics of proteus mirabilis isolated from domestic rats: an environmental driver to antimicrobial resistance in public health in Arusha Tanzania
Authors: Floramanka P. Ndakidemi, Maneno E. Baravuga, Alexanda Mzula and Abdul S. Katakweba
Ger. J. Microbiol.
2023.
vol. 3, Iss. 1
pp:13-23
Doi: https://doi.org/10.51585/gjm.2023.1.0022
Abstract:
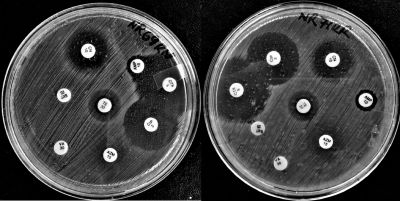
Proteus mirabilis (P. mirabilis) is a bacterial pathogen contributing to opportunistic infections, nosocomial outbreaks, and mostly hematogenous ascending urinary tract infections. It has repeatedly been found in rats. Due to rat-human interaction, rats are likely responsible for spreading these bacteria and their antimicrobial-resistant. This study was performed to genetically characterize and assess antimicrobial susceptibility patterns of P. mirabilis isolated from rats cohabiting with humans in Arusha municipality, Tanzania. A total of 139 rats were trapped from March to May 2021 and identified at the species level using morphological and morphometric features. Deep-intestinal swabs were obtained and pre-enriched in buffered peptone water. P. mirabilis was isolated by conventional culture and biochemical methods and confirmed by 16S rRNA polymerase chain reaction and sequencing. Phylogenetics was used to assess the similarities of the isolates. Antimicrobial susceptibility test was done by disk diffusion method using seven antibiotics, including tetracycline, ciprofloxacin, gentamicin, cefotaxime, trimethoprim-sulfamethoxazole, azithromycin, and ampicillin. Resistance genes blaTEM, tetA, tetB, mphA, blaSHV, blaCTX-M, sul1, and sul2 were traced in each isolate using PCR. Mixed rat species, Rattus rattus (55.4%), Mus musculus (15.8%), and Mastomys natalensis (28.8%), were captured. P. mirabilis was isolated from four (2.9%) Rattus rattus samples. By PCR and sequencing, all were confirmed as P. mirabilis and 100% similar to strains from GenBank. Three isolates showed multidrug resistance (MDR) against trimethoprim-sulfamethoxazole, azithromycin, and ampicillin, while all isolates were resistant to azithromycin and ampicillin, and susceptible to ciprofloxacin, gentamicin, and cefotaxime. Three were resistance to trimethoprim-sulfamethoxazole and intermediate to tetracycline. PCR analysis detected tetA, blaTEM, sul1, and sul2 resistance genes. Constructed phylogenetic tree showed that all isolates from this study were closely related to isolates from Tunisia. The study has discovered the first P. mirabilis isolates from rats in Tanzania with antimicrobial resistance traits that could be of public health concern.
Keywords:
P. mirabilis, Rats, Antimicrobial resistance, Resistance genes, 16S rRNA
Statistics:
Article Views: 1411
PDF Download: 178