WGS analysis of hypervirulent and MDR Klebsiella pneumoniae from Vietnam reveales an inverse relationship between resistome and virulome
Authors: Gamal Wareth, Hanka Brangsch, Ngoc H. Nguyen, Tuan N.M. Nguyen, Mathias W. Pletz, Heinrich Neubauer and Lisa D. Sprague
Ger. J. Microbiol.
2024.
vol. 4, Iss. 1
pp:15-24
Doi: https://doi.org/10.51585/gjm.2024.1.0030
Abstract:
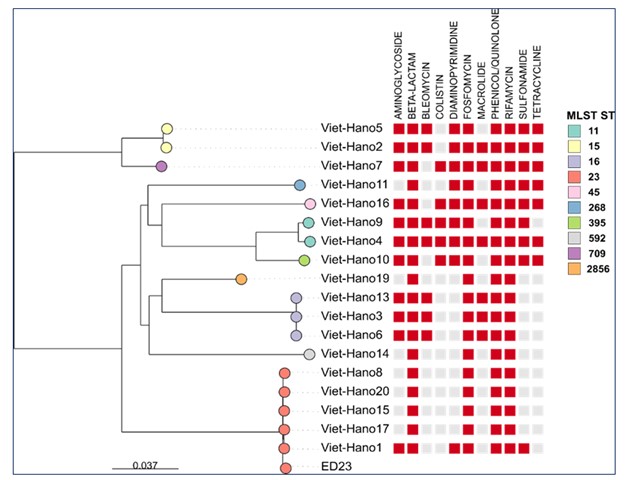
The emergence of Klebsiella (K.) pneumoniae as a leading cause of nosocomial infections in Southeast Asia is of concern. Vietnam has an outstanding position with regard to antimicrobial-resistant (AMR) pathogens and is a hotspot for carbapenem-resistant K. pneumoniae. In the current study, 19 clinical K. pneumoniae strains isolated from patients in Vietnam were tested for antibiotic susceptibility, and their genome sequences were analyzed to investigate potential resistance profiles, genotypes, AMR determinants, and virulence-associated genes. More than half of the isolates were multidrug-resistant (MDR), displaying resistance to ciprofloxacin, levofloxacin, chloramphenicol, piperacillin, cefotaxime, ceftazidime, and cefepime. Carbapenem- resistance was detected in 47% (n=9) of isolates. Five isolates were assigned to sequence type (ST) 23 by multi-locus sequence typing (MLST). These ST23 strains exhibited determinants characteristic of hypervirulent strains and were sensitive to all antibiotics tested except for one strain, which was resistant to chloramphenicol and trimethoprim/sulfamethoxazole. All isolates had AMR determinants that conferred resistance to aminoglycosides and ß-lactams. Single nucleotide variants of oqxA and oqxB conferring resistance to phenicol/quinolone, ompK37 and ompA conferring resistance to beta-lactams, and lptD conferring resistance to rifamycin, were found in all isolates. Additionally, 95% of the isolates (n=18) carried fosA genes; however, only two were resistant to fosfomycin. Five isolates carried genes conferring resistance to colistin; only one carried mcr-1.1 and showed resistance to colistin. One hundred thirty virulence-associated genes were identified. The current study demonstrated an inverse relationship between the number of detected virulence determinants and antibiotic resistance. Several well-known resistance genes have been identified but did not mediate resistance, which could be due to insufficient levels or lack of expression. A comprehensive study of the genotypic findings of AMR determinants and virulence with phenotypic data is required. The presence of hypervirulent K. pneumoniae carrying specific virulomes is a growing problem, and monitoring of virulence characteristics of K. pneumoniae is important.
Keywords:
Klebsiella pneumoniae, Phenotyping, Hypervirulent MDR, WGS, Resistome, Plasmidome, Virulome, Vietnam
Statistics:
Article Views: 174
PDF Download: 15